Chapter 3 Using ggVennDiagram
Generate example data.
genes <- paste0("gene",1:1000)
set.seed(20210302)
gene_list <- list(A = sample(genes,100),
B = sample(genes,200),
C = sample(genes,300),
D = sample(genes,200))
library(ggVennDiagram)3.1 long category names
If you use long category names in Venn plot, labels may be cropped by plot borders. To avoid this, just use a ggplot trick to expand x axis.
p1 <- ggVennDiagram(gene_list,
category.names = c("a very long name","short name","name","another name"))
p1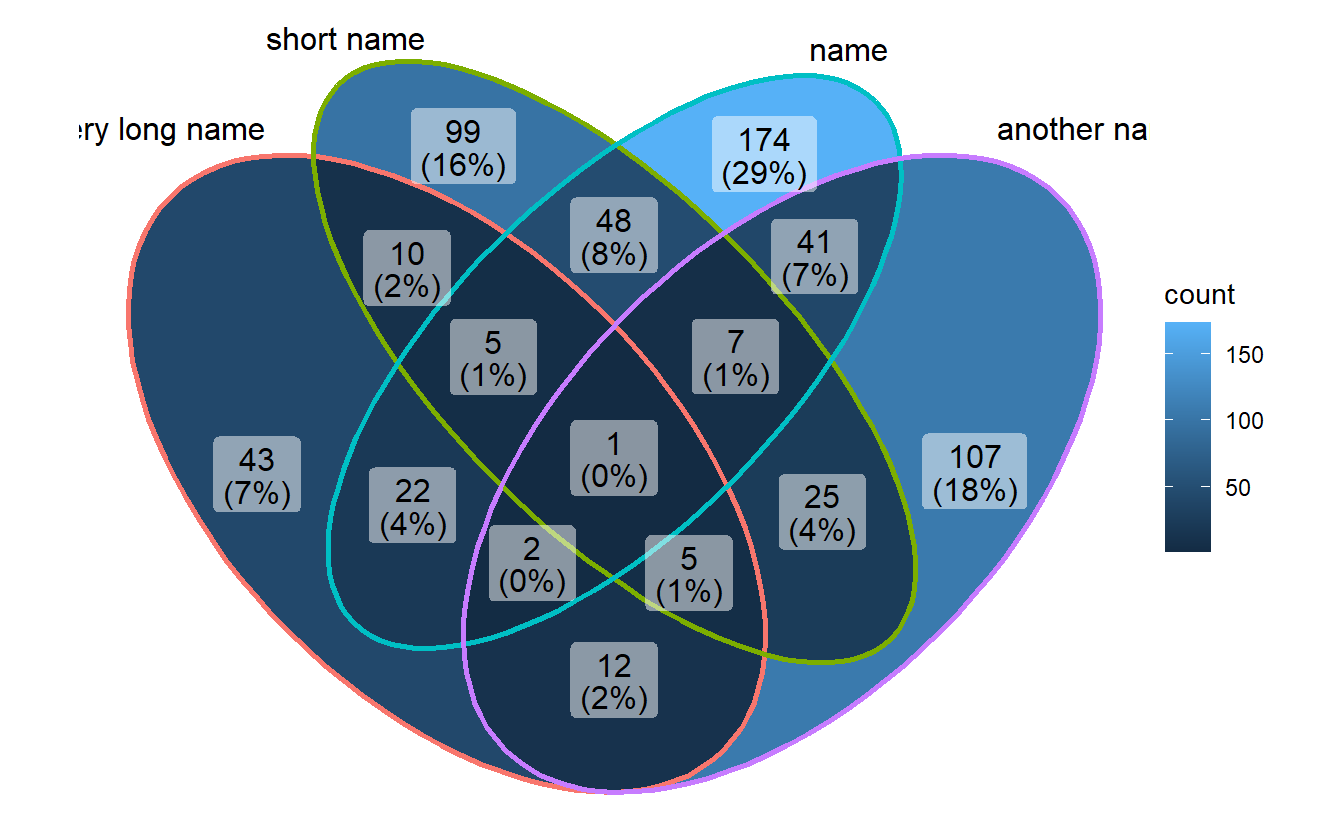
# expand axis to show long set labels
p1 + scale_x_continuous(expand = expansion(mult = .2))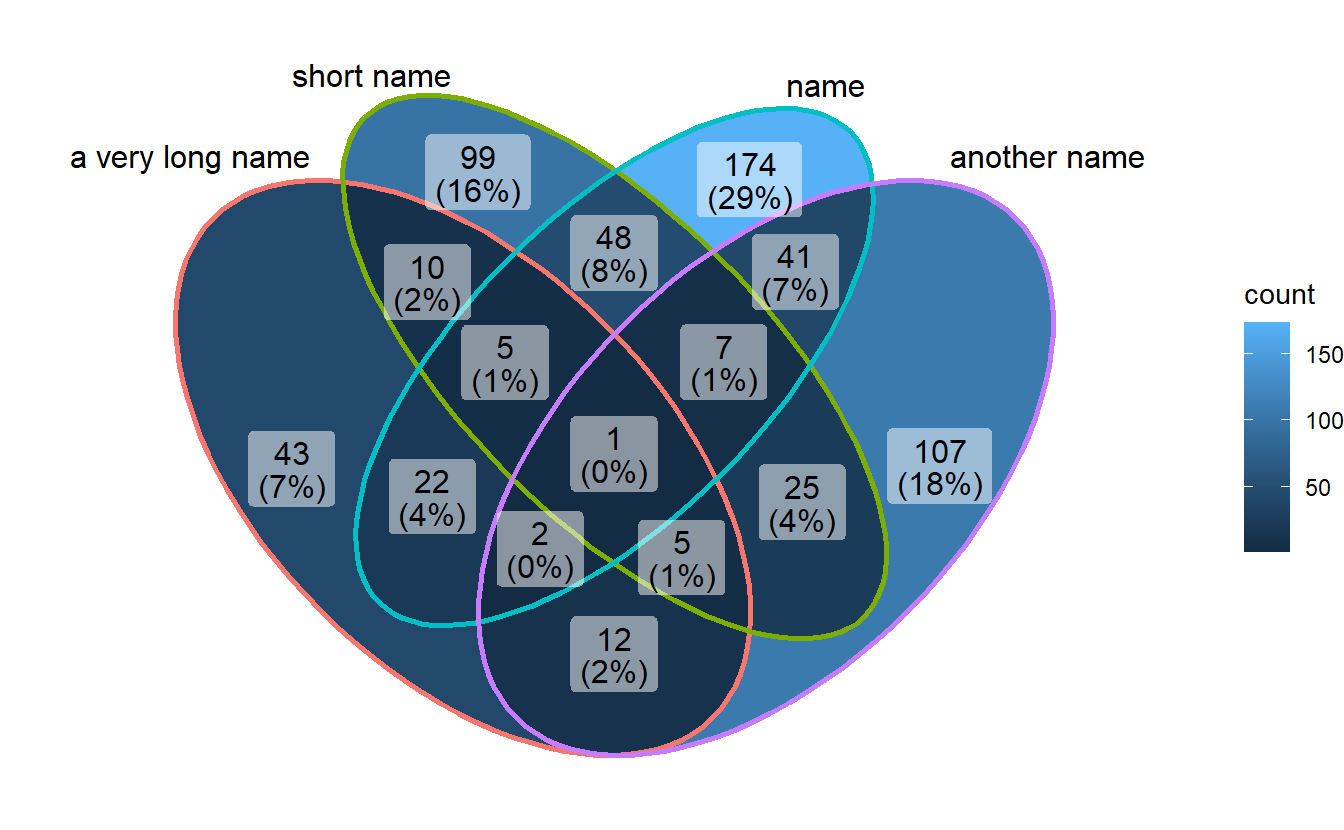
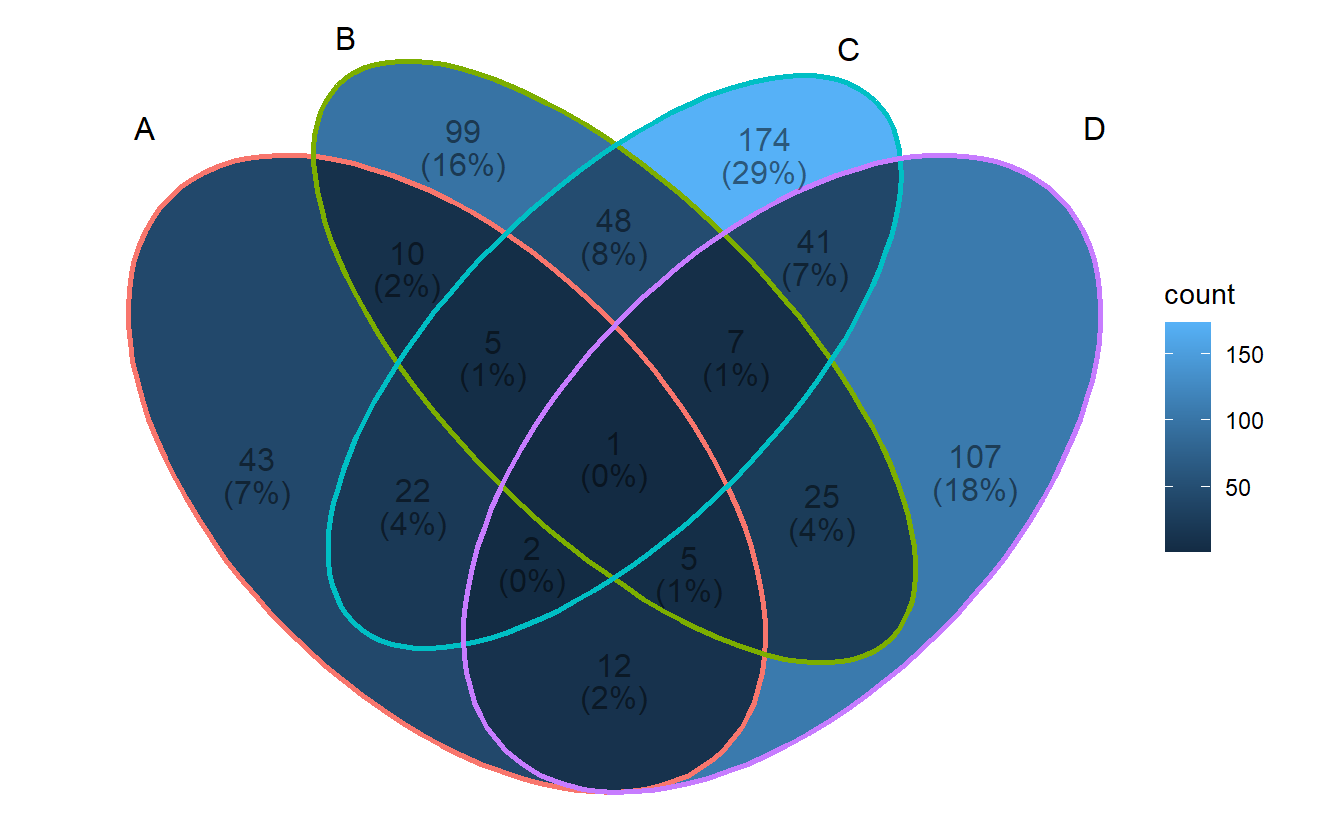
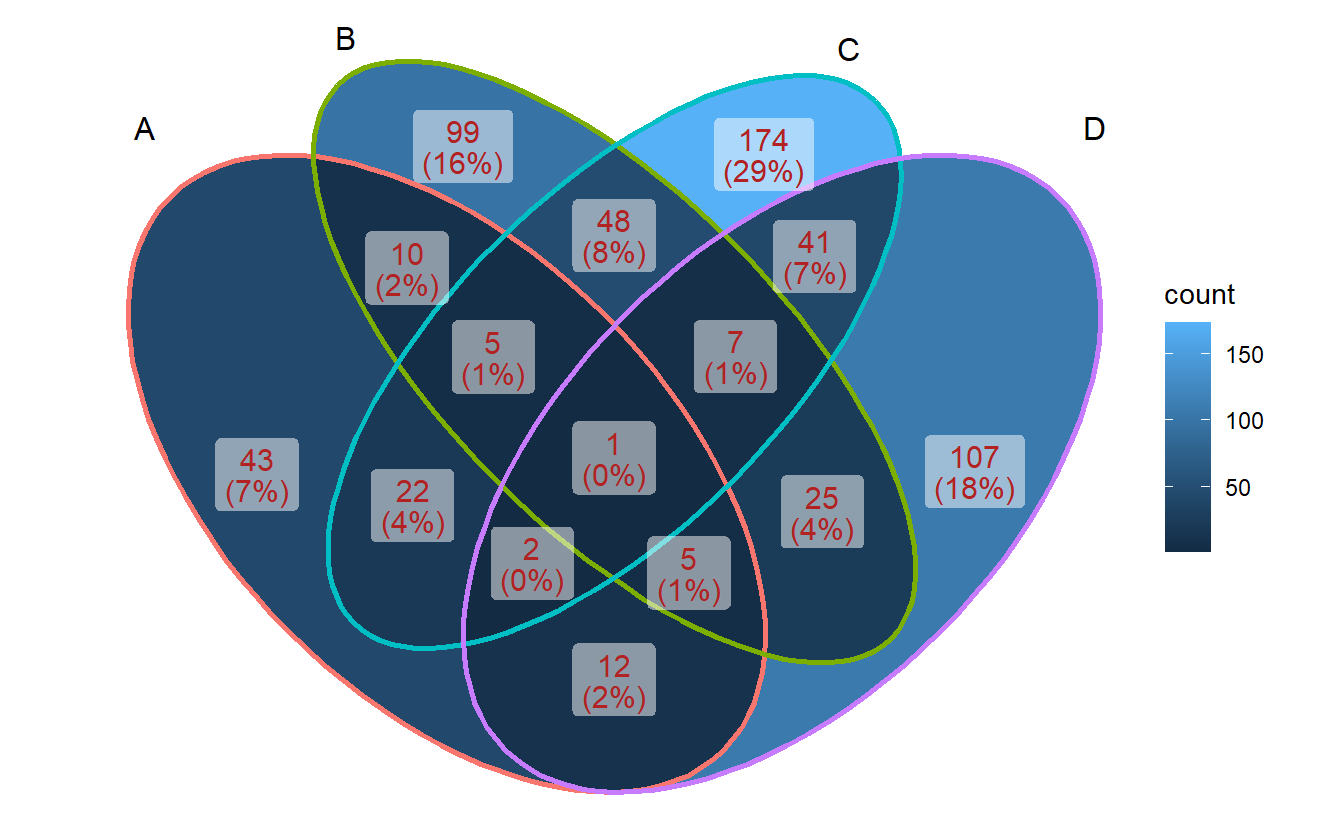
3.2 Show intersection values
When intersection values only have several members, ggVennDiagram is efficient to show the values in places.
set.seed(0)
small_list <- lapply(sample(6:10, size = 4), function(x){
sample(letters,x)
})
ggVennDiagram(small_list,
category.names = LETTERS[1:4],
show_intersect = TRUE)## Warning: Ignoring unknown aesthetics: text3.4 Setting region label
3.4.1 text content
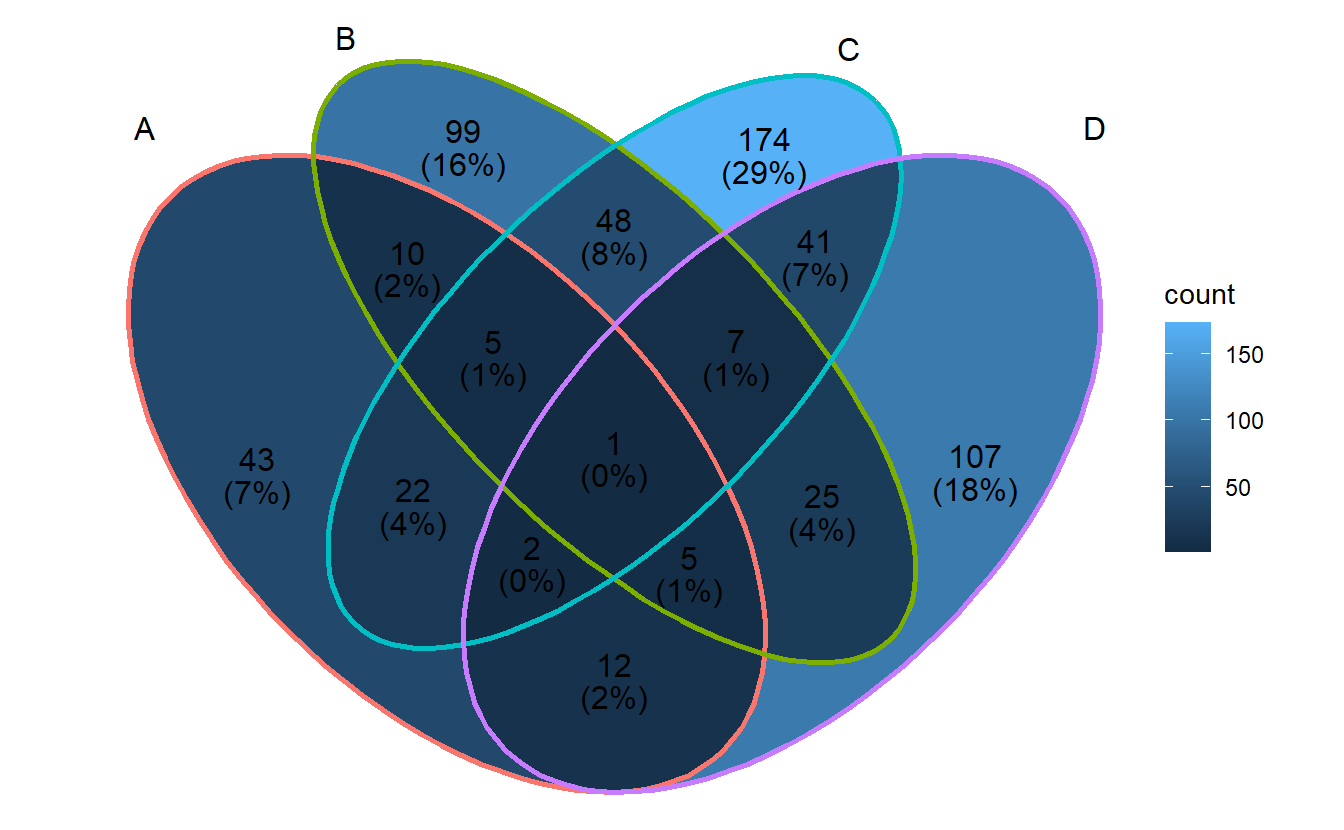
ggVennDiagram(gene_list, label = "count")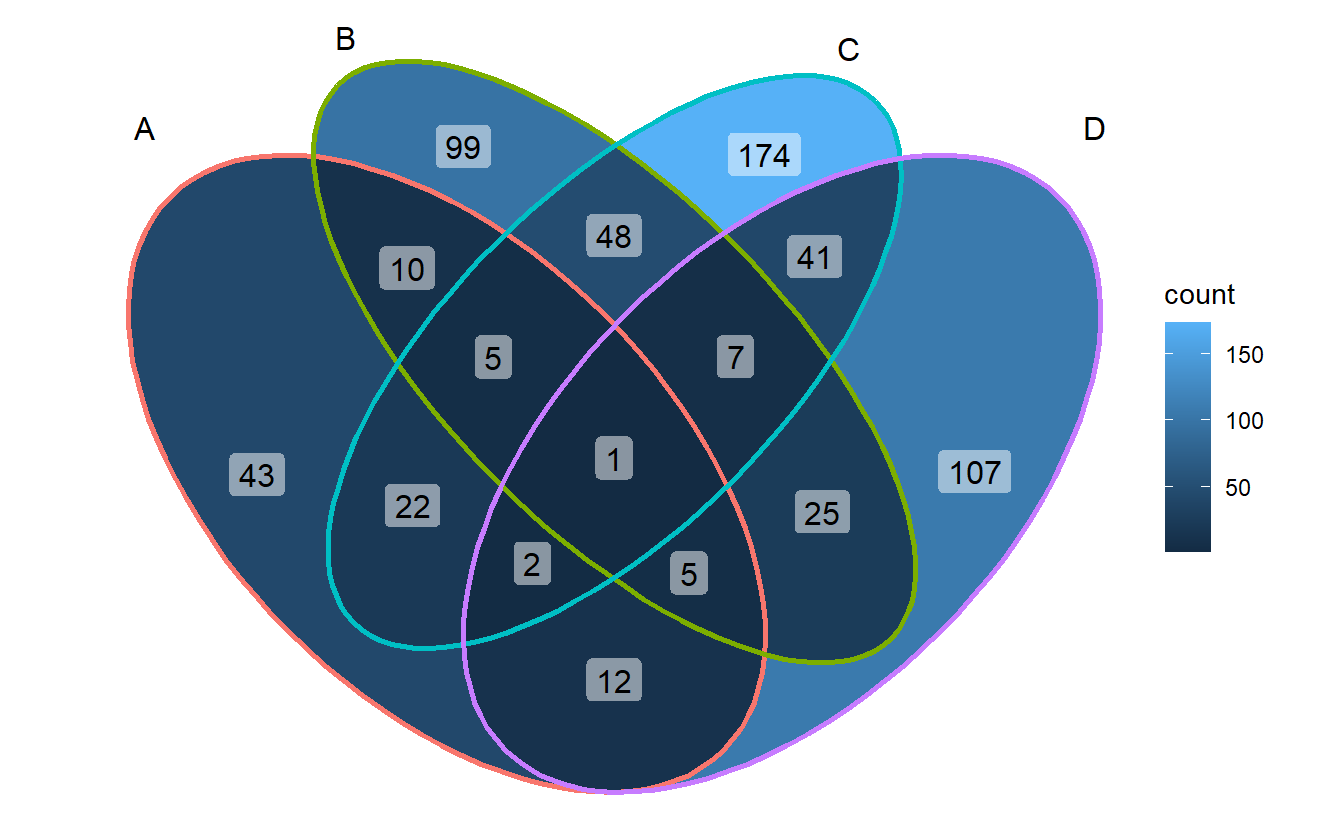
ggVennDiagram(gene_list, label = "percent")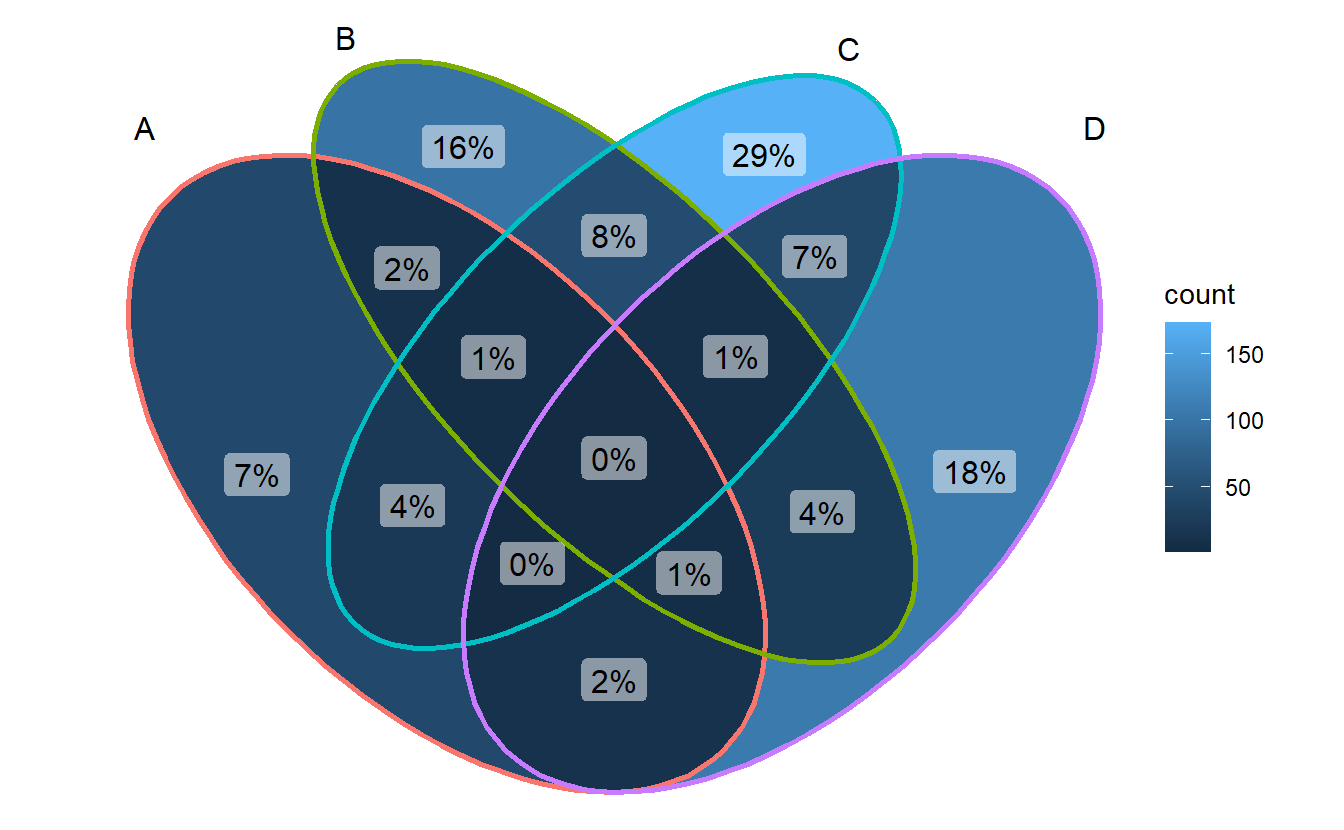
3.6 Changing palette
- changing fill palette
library(ggplot2)
p <- ggVennDiagram(gene_list)
# Red Blue
p + scale_fill_distiller(palette = "RdBu")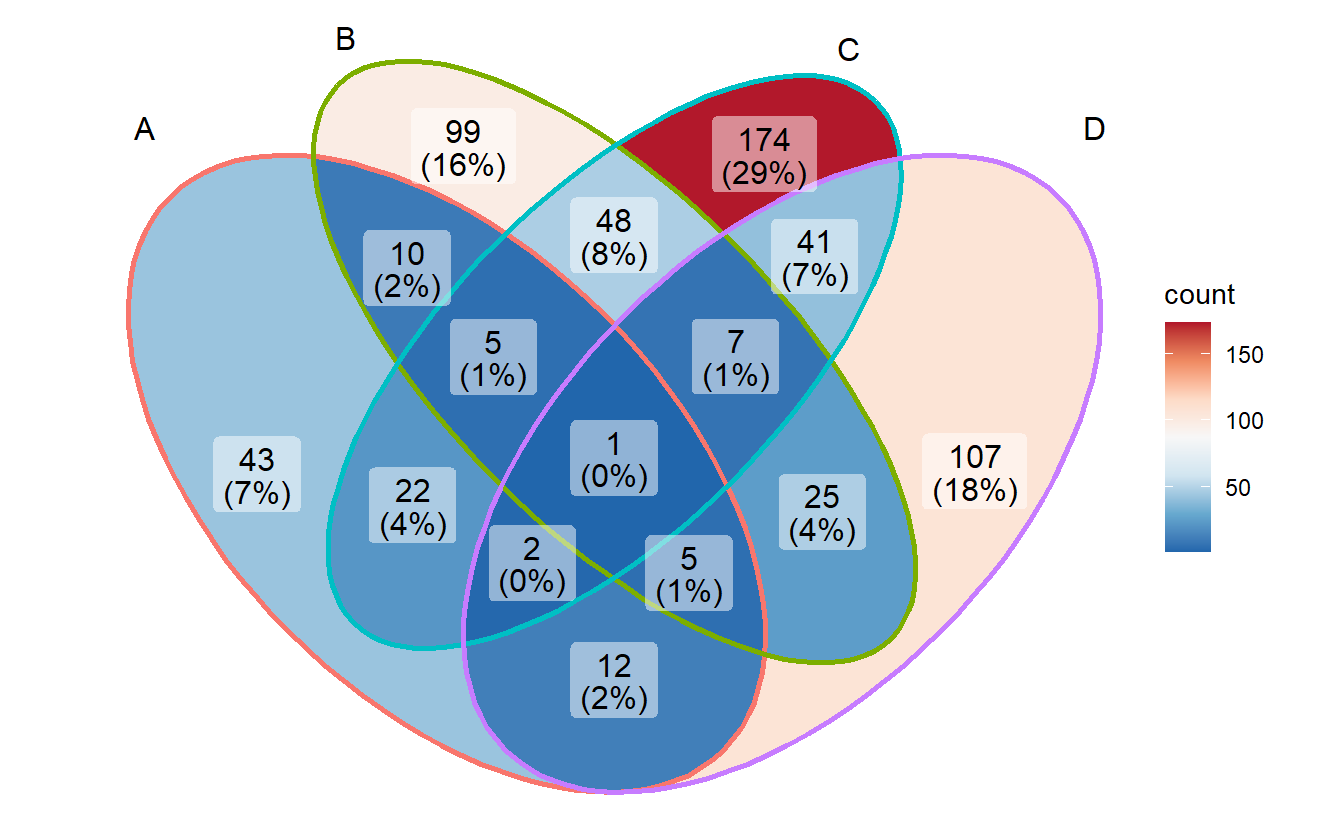
# Reds
p + scale_fill_distiller(palette = "Reds", direction = 1)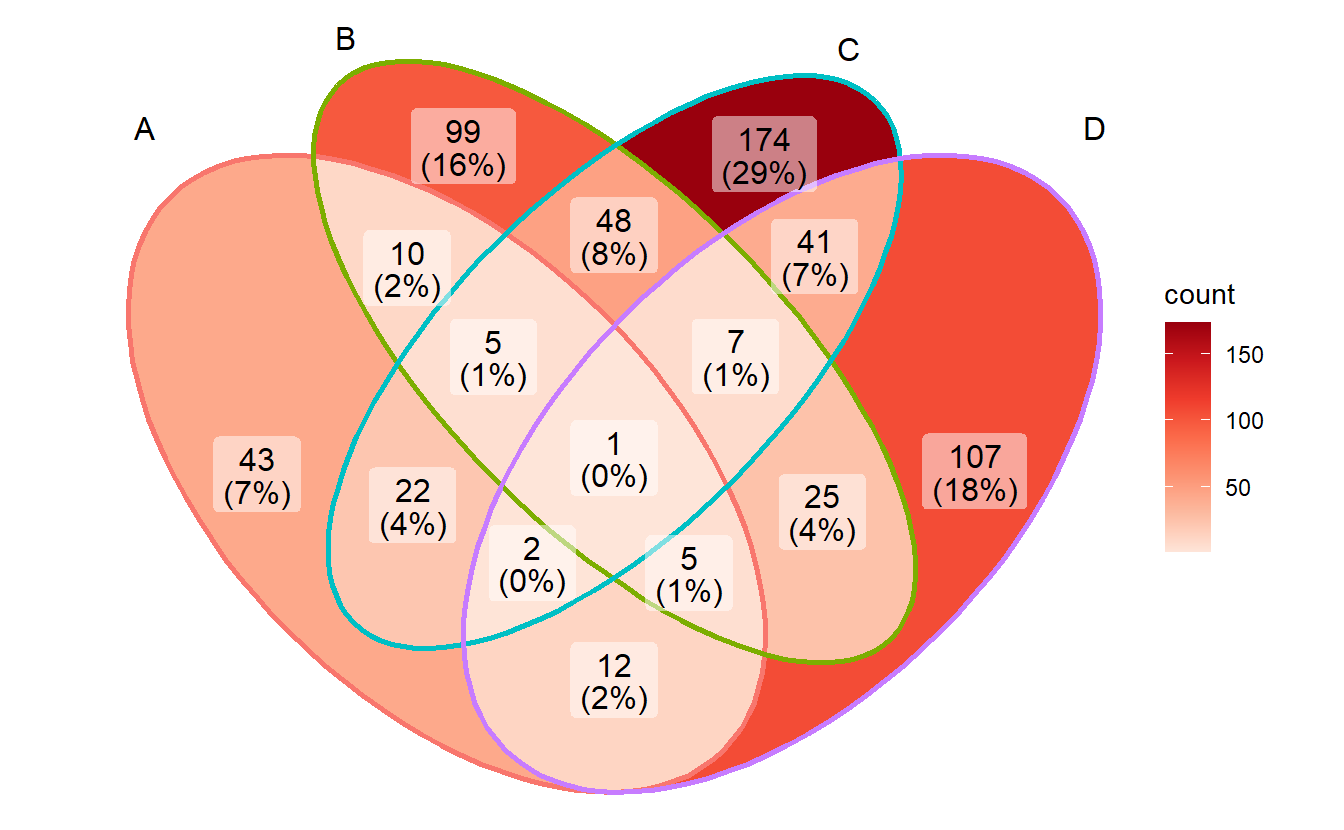
- change edge colors
p + scale_color_brewer(palette = "Set1")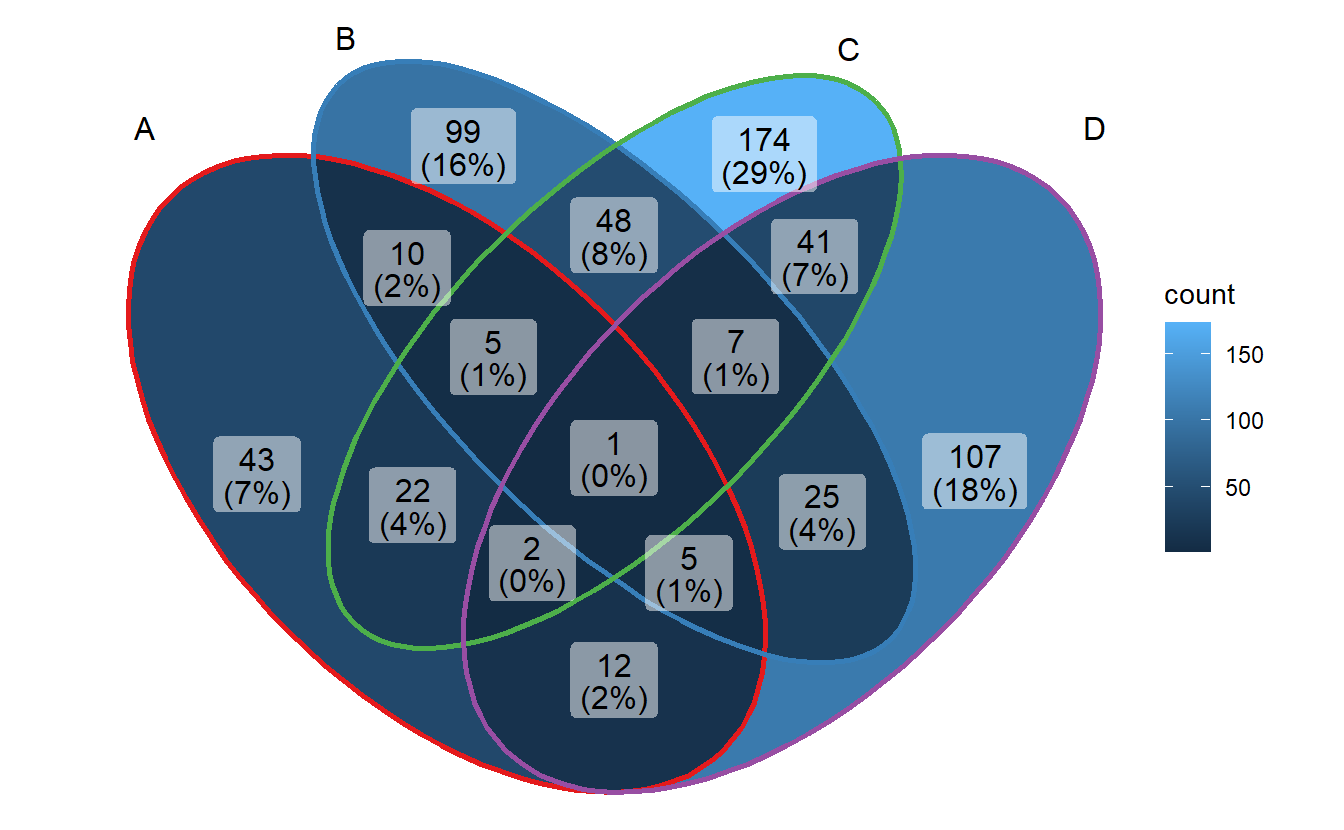
Some other palletes are:
RColorBrewer::display.brewer.all()
3.7 Adding note
p + labs(title = "Four sets Venn Diagram",
subtitle = "generated by `ggVennDiagram`",
caption = Sys.Date())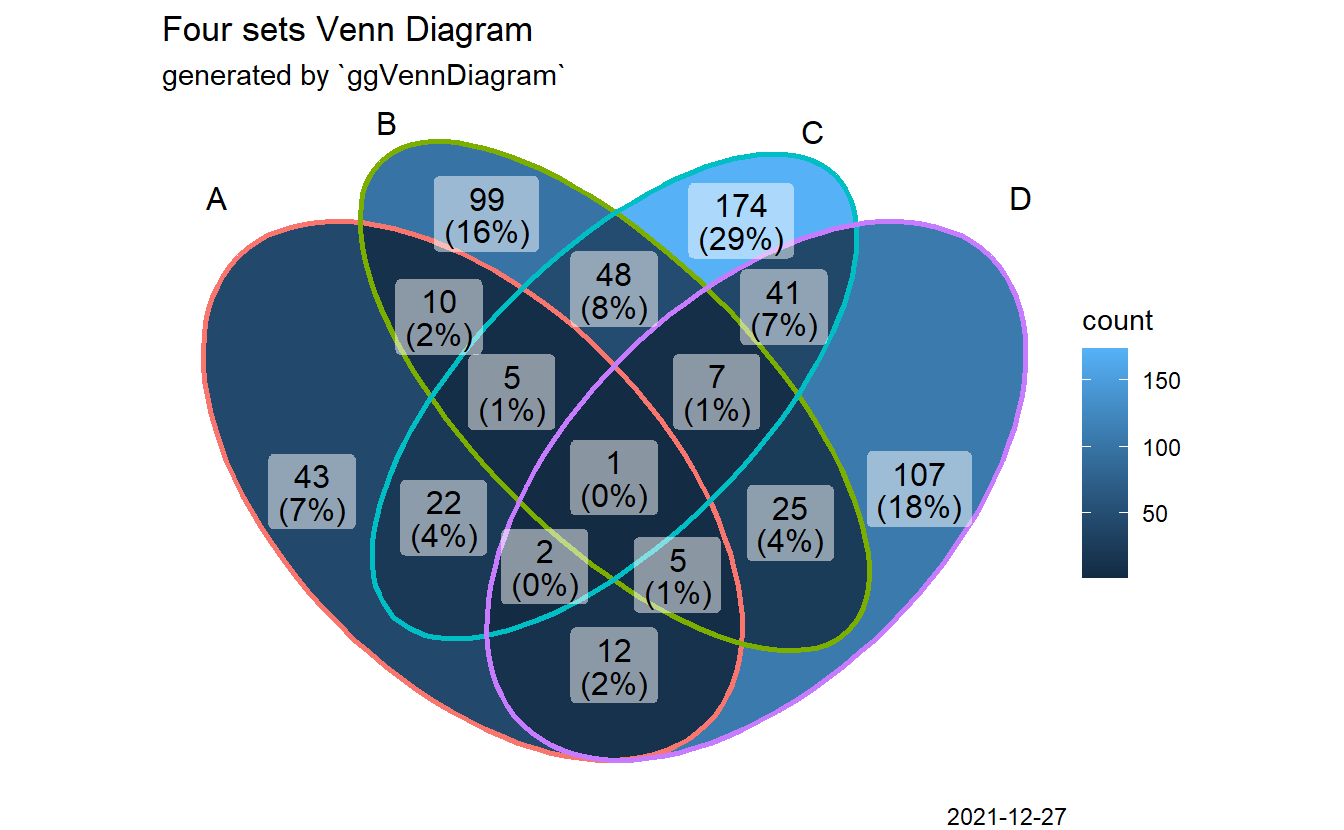
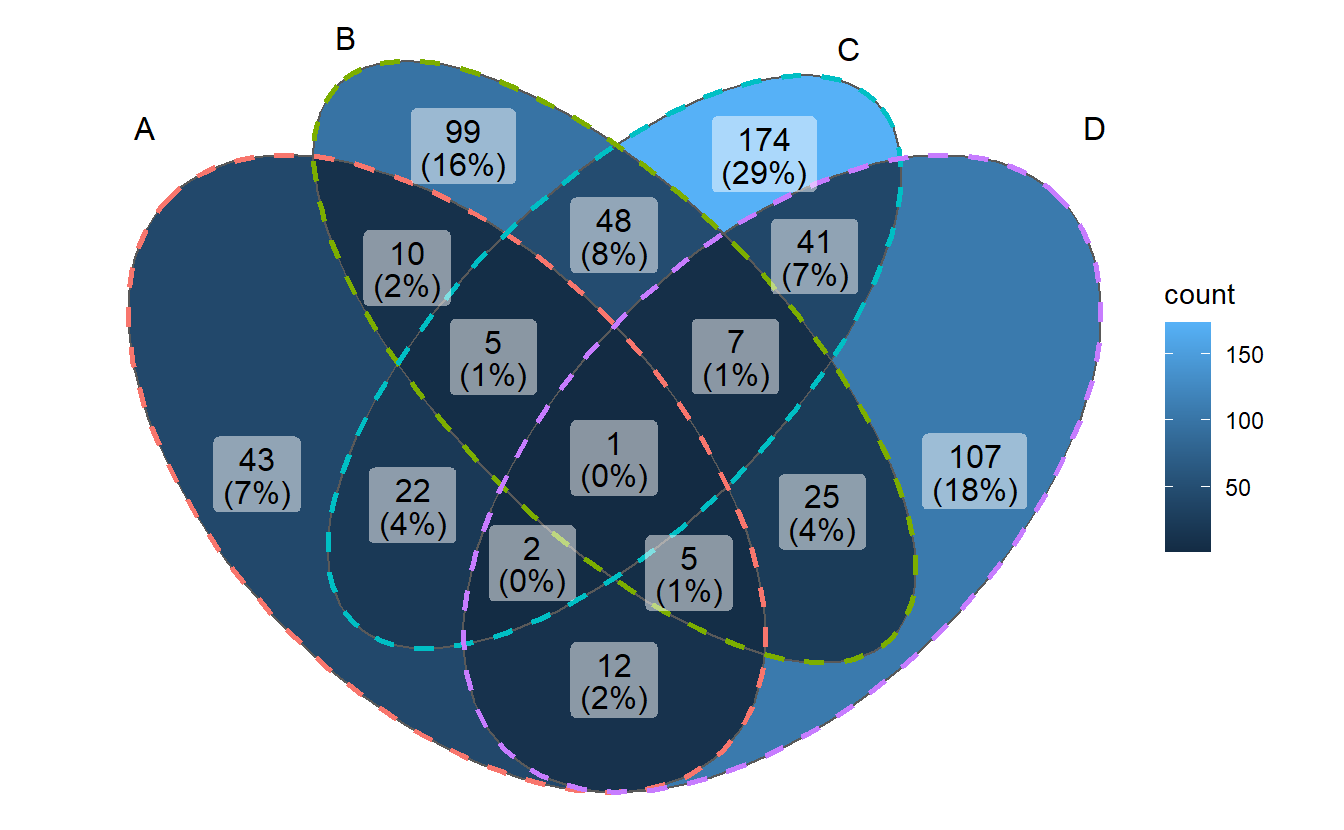
3.8 Comprehensive customization by using helper functions
The main function ggVennDiagram() accepts a list input, and output a ggplot object.
By measuring the length of input list, it automatically applies internal functions
to build a plot in two steps: data preparation and visualization.
Data preparation was packaged into one function process_data(). Its output
is a S4 VennPlotData class object, which contains three slots, setEdge,
setLabel and region. These slot data then can be further plotted with ggplot
functions.
See below for a better understanding.
venn <- Venn(gene_list)
data <- process_data(venn)
ggplot() +
# 1. region count layer
geom_sf(aes(fill = count), data = venn_region(data)) +
# 2. set edge layer
geom_sf(aes(color = id), data = venn_setedge(data), show.legend = FALSE) +
# 3. set label layer
geom_sf_text(aes(label = name), data = venn_setlabel(data)) +
# 4. region label layer
geom_sf_label(aes(label = count), data = venn_region(data)) +
theme_void()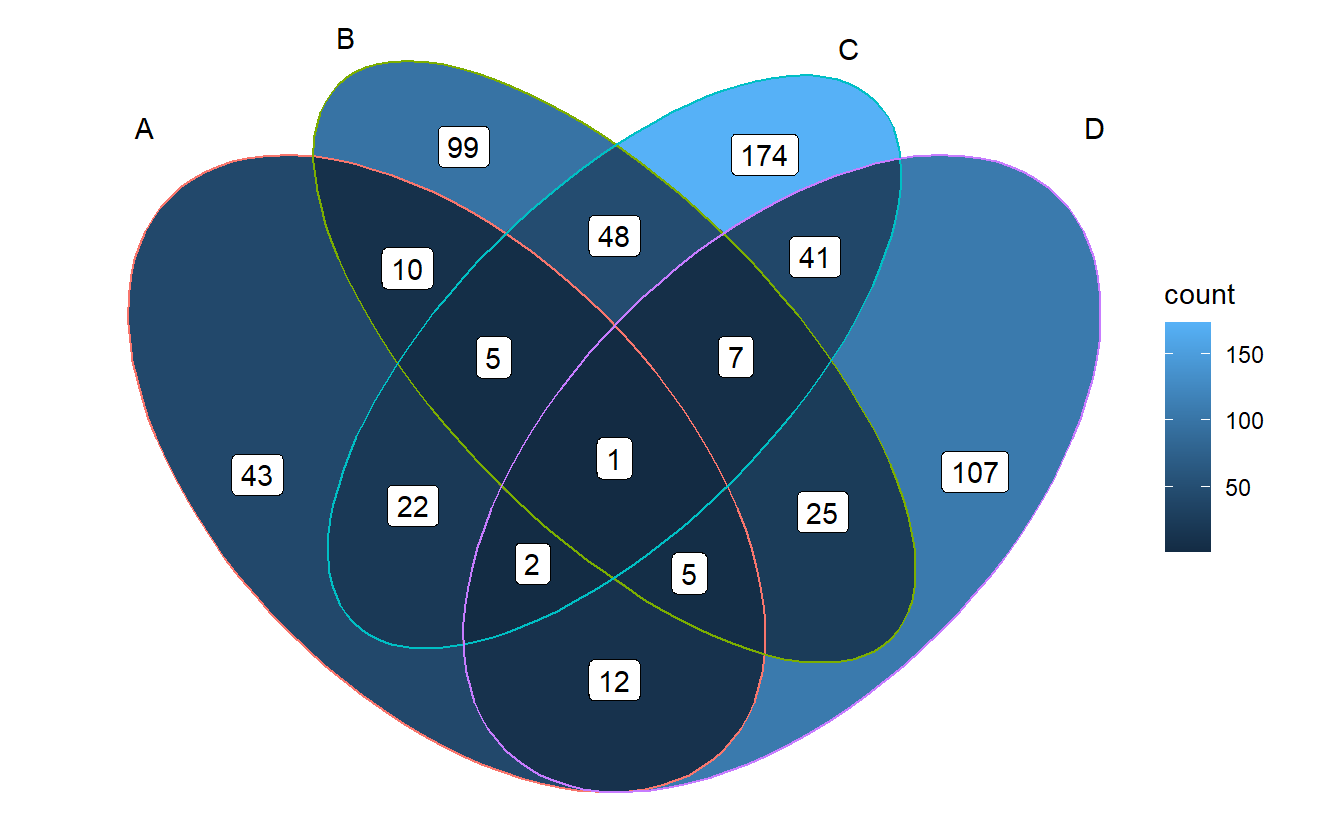
ggVennDiagram export functions to get these data, and they can be used
for comprehensive customization in user-side.
Venn(): Venn object constructor, use this to construct a Venn object from list.process_data(): process data with a Venn objectvenn_region(): get region data to plotvenn_setedge(): get setedge data to plotvenn_setlabel(): get setlabel data to plot
For example, you may change edge/fill/label properties as you will.
ggplot() +
# change mapping of color filling
geom_sf(aes(fill = id), data = venn_region(data), show.legend = FALSE) +
# adjust edge size and color
geom_sf(color="grey", size = 3, data = venn_setedge(data), show.legend = FALSE) +
# show set label in bold
geom_sf_text(aes(label = name), fontface = "bold", data = venn_setlabel(data)) +
# add a alternative region name
geom_sf_label(aes(label = name), data = venn_region(data), alpha = 0.5) +
theme_void()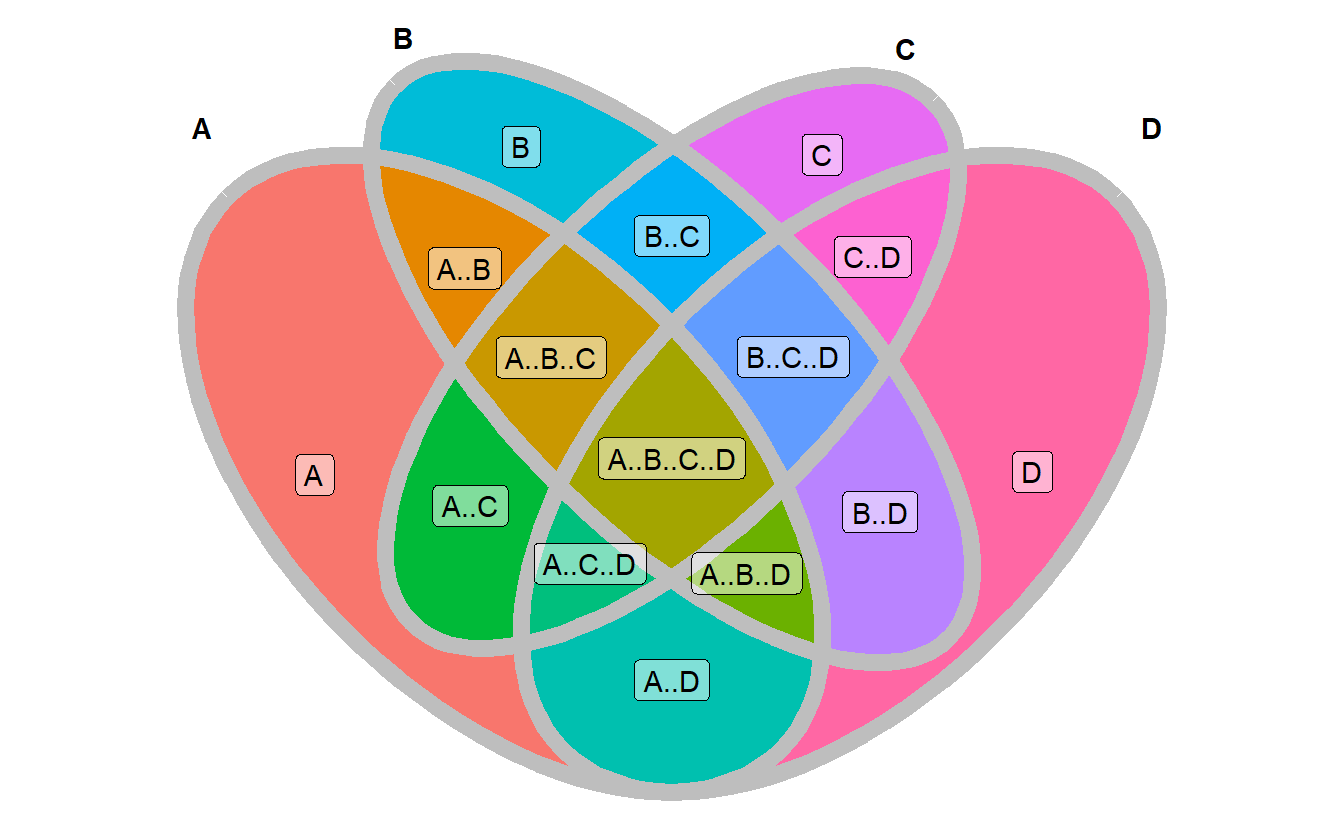
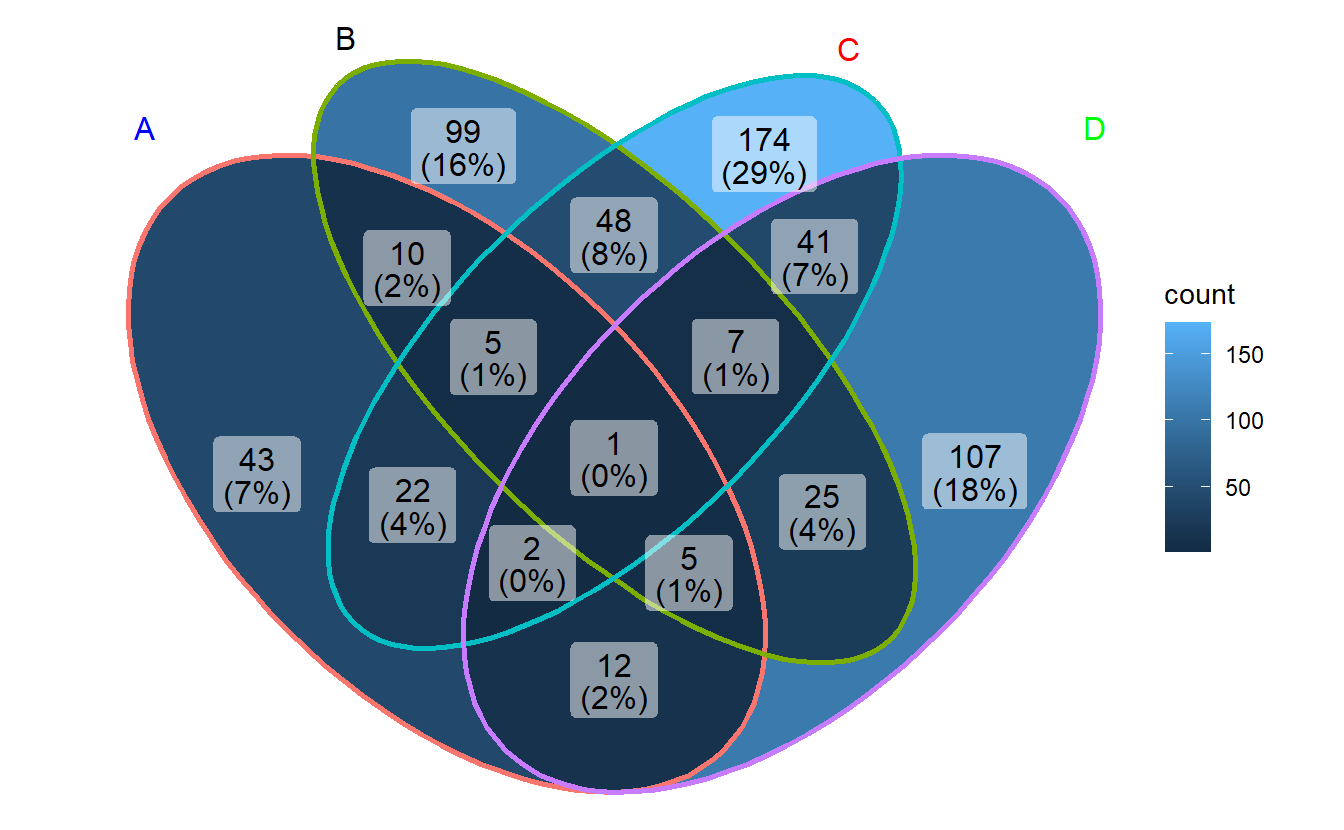
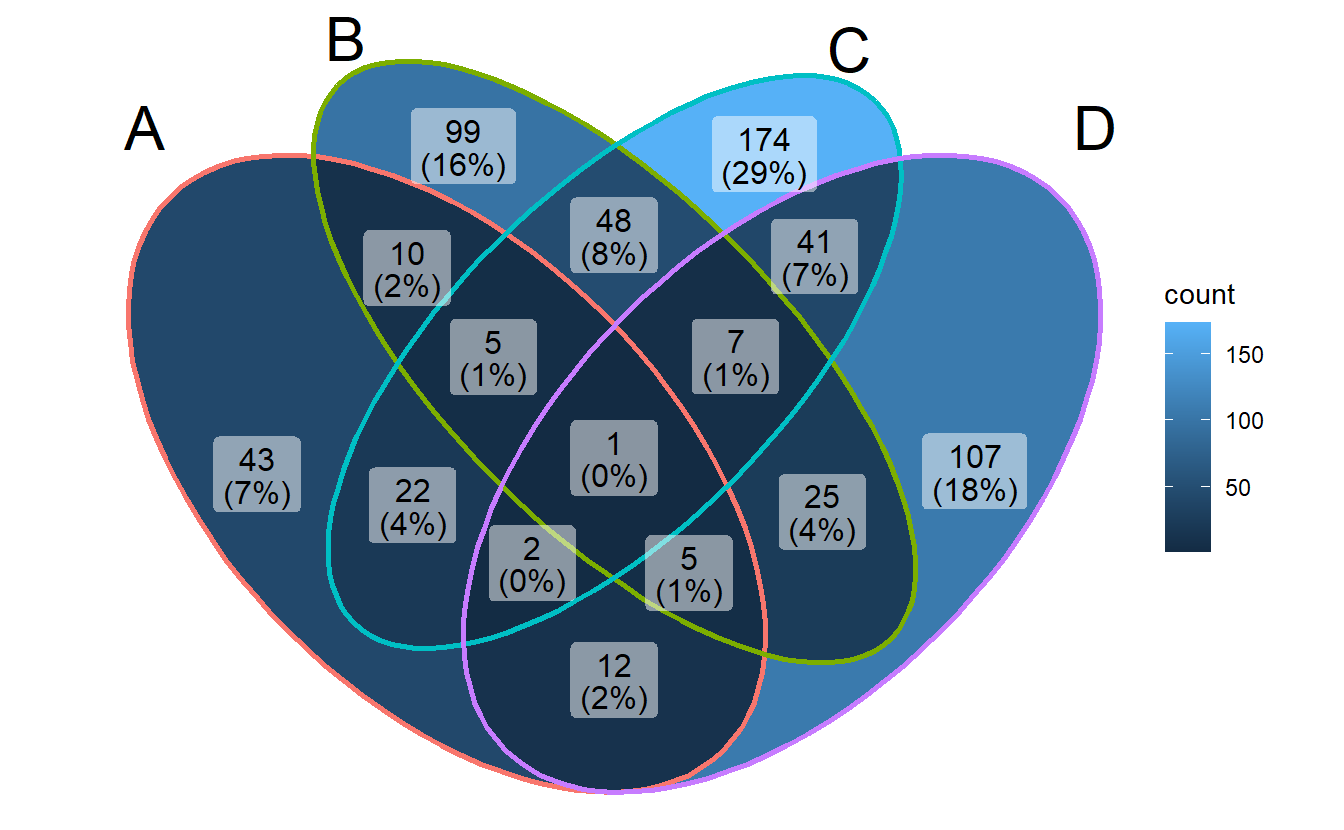
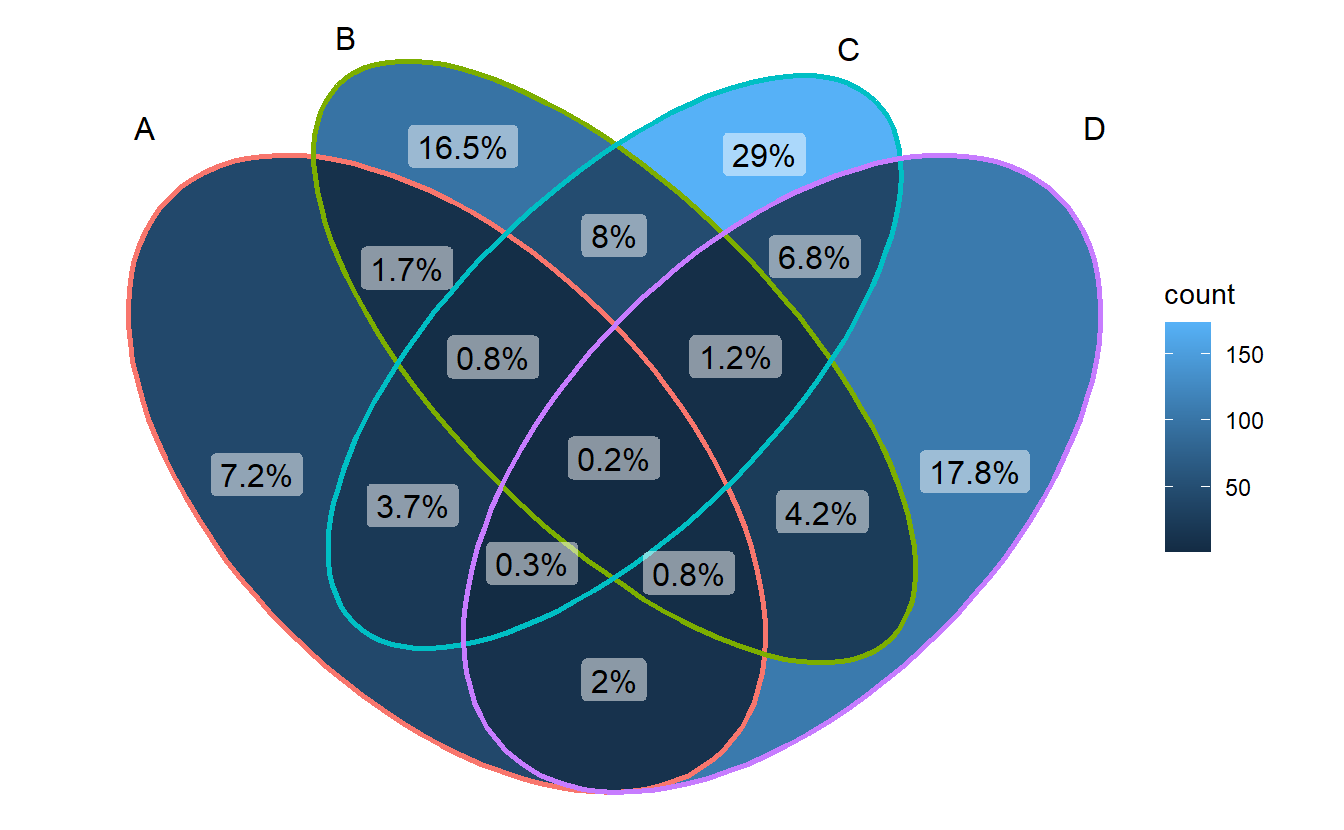
 - Method 2: use
- Method 2: use